Real-world data often has missing values. Data can have missing values for a number of reasons such as observations that were not recorded and data corruption.
Handling missing data is important as many machine learning algorithms do not support data with missing values.
In this tutorial, you will discover how to handle missing data for machine learning with Python.
Specifically, after completing this tutorial you will know:
- How to mark invalid or corrupt values as missing in your dataset.
- How to confirm that the presence of marked missing values causes problems for learning algorithms.
- How to remove rows with missing data from your dataset and evaluate a learning algorithm on the transformed dataset.
This tutorial is divided into 4 parts; they are:
- Diabetes Dataset
- Mark Missing Value
- Missing Value Cause Problems
- Remove Row With Missing Values
1. Diabetes Dataset
As the basis of this tutorial, we will use the so-called diabetes dataset that has been widely studied as a machine learning dataset since the 1990s.
The dataset classifies patient data as either an onset of diabetes within five years or not. There are 768 examples and 8 input variables.
We will aim for this region, but note that the models in this tutorial are not optimized; they are designed to demonstrate feature selection schemes.
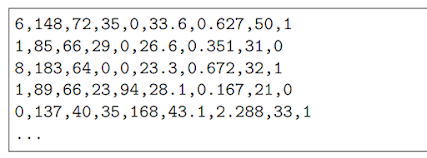 |
| Example of rows that contains missing value
|
There are missing observations for some columns that are marked as a zero value.
We can corroborate this by the definition of those columns and the domain knowledge that a zero value is invalid for those measures, e.g. a zero for body mass index or blood pressure is invalid.
2. Mark Missing Values
Most data has missing values, and the likelihood of having missing values increases with the size of the dataset.
In this section, we will look at how we can identify and mark values as missing. We can use plots and summary statistics to help identify missing or corrupt data.
# load and summarize the dataset
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# summarize the dataset
print(dataset.describe())
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# summarize the dataset
print(dataset.describe())
-----Result-----
 |
| Example output from calculating summary statistics for each variable
|
We can see that there are columns that have a minimum value of zero (0). On some columns, a value of zero does not make sense and indicates an invalid or missing value.
Let’s confirm this by looking at the raw data, the example prints the first 20 rows of data.
# load the dataset and review rows
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# summarize the first 20 rows of data
print(dataset.head(20))
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# summarize the first 20 rows of data
print(dataset.head(20))
-----Result-----
We can get a count of the number of missing values on each of these columns.
# example of summarizing the number of missing values for each variable
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# count the number of missing values for each column
num_missing = (dataset[[1,2,3,4,5]] == 0).sum()
# report the results
print(num_missing)
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# count the number of missing values for each column
num_missing = (dataset[[1,2,3,4,5]] == 0).sum()
# report the results
print(num_missing)
-----Result-----
1 5
2 35
3 227
4 374
5 11
2 35
3 227
4 374
5 11
In Python, specifically Pandas, NumPy and Scikit-Learn, we mark missing values as NaN. Values with a NaN value are ignored from operations like sum, count, etc.
We can mark values as NaN easily with the Pandas DataFrame by using the replace() function on a subset of the columns we are interested in. After we have marked the missing values, we can use the isnull() function to mark all of the NaN values in the dataset as True and get a count of the missing values for each column.
# example of marking missing values with nan values
from numpy import nan
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# count the number of nan values in each column
print(dataset.isnull().sum())
from numpy import nan
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# count the number of nan values in each column
print(dataset.isnull().sum())
-----Result-----
0 0
1 5
2 35
3 227
4 374
5 11
6 0
7 0
8 0
dtype: int64
1 5
2 35
3 227
4 374
5 11
6 0
7 0
8 0
dtype: int64
# example of review data with missing values marked with a nan
from numpy import nan
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# summarize the first 20 rows of data
print(dataset.head(20))
from numpy import nan
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# summarize the first 20 rows of data
print(dataset.head(20))
-----Result-----
3. Missing Values Cause Problems
Having missing values in a dataset can cause errors with some machine learning algorithms.
In this section, we will try to evaluate the Linear Discriminant Analysis (LDA) algorithm on the dataset with missing values. This is an algorithm that does not work when there are missing values in the dataset.
# example where missing values cause errors
from numpy import nan
from pandas import read_csv
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# split dataset into inputs and outputs
values = dataset.values
X = values[:,0:8]
y = values[:,8]
# define the model
model = LinearDiscriminantAnalysis()
# define the model evaluation procedure
cv = KFold(n_splits=3, shuffle=True, random_state=1)
# evaluate the model
from numpy import nan
from pandas import read_csv
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# split dataset into inputs and outputs
values = dataset.values
X = values[:,0:8]
y = values[:,8]
# define the model
model = LinearDiscriminantAnalysis()
# define the model evaluation procedure
cv = KFold(n_splits=3, shuffle=True, random_state=1)
# evaluate the model
result = cross_val_score(model, X, y, cv=cv, scoring='accuracy')
# report the mean performance
print('Accuracy: %.3f' % result.mean())
# report the mean performance
print('Accuracy: %.3f' % result.mean())
-----Result-----
ValueError: Input contains NaN, infinity or a value too large for dtype('float64').
4. Remove Rows With Missing Values
The simplest strategy for handling missing data is to remove records that contain a missing value.
Pandas provides the dropna() function that can be used to drop either columns or rows with missing data.
# example of removing rows that contain missing values
from numpy import nan
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# summarize the shape of the raw data
print(dataset.shape)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# drop rows with missing values
dataset.dropna(inplace=True)
# summarize the shape of the data with missing rows removed
print(dataset.shape)
from numpy import nan
from pandas import read_csv
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# summarize the shape of the raw data
print(dataset.shape)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# drop rows with missing values
dataset.dropna(inplace=True)
# summarize the shape of the data with missing rows removed
print(dataset.shape)
-----Result-----
(768, 9)
(392, 9)
(392, 9)
We now have a dataset that we could use to evaluate an algorithm sensitive to missing values like LDA.
# evaluate model on data after rows with missing data are removed
from numpy import nan
from pandas import read_csv
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# drop rows with missing values
dataset.dropna(inplace=True)
# split dataset into inputs and outputs
values = dataset.values
X = values[:,0:8]
y = values[:,8]
# define the model
model = LinearDiscriminantAnalysis()
# define the model evaluation procedure
cv = KFold(n_splits=3, shuffle=True, random_state=1)
# evaluate the model
result = cross_val_score(model, X, y, cv=cv, scoring='accuracy')
# report the mean performance
print('Accuracy: %.3f' % result.mean())
from numpy import nan
from pandas import read_csv
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
# load the dataset
dataset = read_csv('pima-indians-diabetes.csv', header=None)
# replace '0' values with 'nan'
dataset[[1,2,3,4,5]] = dataset[[1,2,3,4,5]].replace(0, nan)
# drop rows with missing values
dataset.dropna(inplace=True)
# split dataset into inputs and outputs
values = dataset.values
X = values[:,0:8]
y = values[:,8]
# define the model
model = LinearDiscriminantAnalysis()
# define the model evaluation procedure
cv = KFold(n_splits=3, shuffle=True, random_state=1)
# evaluate the model
result = cross_val_score(model, X, y, cv=cv, scoring='accuracy')
# report the mean performance
print('Accuracy: %.3f' % result.mean())
-----Result-----
Accuracy: 0.781


No comments:
Post a Comment